
MICROORGANISMS
Microorganisms play profound roles in pet health. Trillions of microbes in the intestinal tract create molecular networks with body-wide impacts, including effects on nutrient digestibility, gut health, brain health, immune function and longevity.
Historically, researchers studied microbes one species or strain at a time. With metagenomics, Purina scientists employ advanced DNA sequencing technologies that simultaneously study thousands of microorganisms to gain greater understanding of the complex interactions that affect health.
Metagenomics show that microbes may rival genomes for their impact on health. Unlike genomes, however, microbiomes shift with dietary changes. In the future, managing microbes with nutrition will optimize not only gut health, but overall physical and mental health, too.
Purina's research

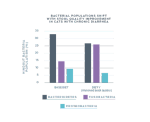
In one study, researchers extracted DNA from the stool of cats with chronic diarrhea to identify bacterial populations in the lower intestinal microbiome.1
Improved stool quality correlated with microbial population changes.
Cats started the clinical trial with chronic diarrhea and a fecal socre of 6 or 7. As the diet trials progressed, almost 50% of the cats experienced improved stool quality, receiving a lower score of 3 on the fecal chart.1,2
As the cats’ chronic diarrhea improved during the study, with dietary changes, three predominant bacterial populations also made significant shifts. These results suggest these bacteria may be important for gut health.

Key things to remember
- Gut microbes may rival the genome for their impact on health.
- Metagenomic approaches allow scientists to study thousands of gut microbes simultaneously to gain a better understanding of how they may impact health and disease.
- The microbial population of the gut can shift with disease and dietary changes, providing an opportunity to improve health with nutritional interventions.
Find out more
1. Ramadan, Z., Xu, H., Laflamme, D., Czarnecki-Maulden, G., Li, Q. J., Labuda, J., & Bourqui, B. (2014). Fecal microbiota of cats with naturally occurring chronic diarrhea assessed using 16S rRNA gene 454-pyrosequencing before and after dietary treatment. Journal of Veterinary Internal Medicine, 28(1), 59–65.
2. Laflamme, D. P., Xu, H., Cupp, C. J., Kerr, W. W., Ramadan, Z., & Long, G. M. (2012). Evaluation of canned therapeutic diets for the management of cats with naturally occurring chronic diarrhea. Journal of Feline Medicine and Surgery, 14, 669–677.